In preparation for training with irregular time indices, with simulated data data will be adaptively downsampled with RDP.
Running RDP on the datasets
"~/cernbox/hysteresis/dipole/datasets/simulation/train_piecewise_eddy_24h.parquet"
"~/cernbox/hysteresis/dipole/datasets/simulation/train_piecewise_eddy_1h.parquet"
"~/cernbox/hysteresis/calibration_fn/SPS_MB_I2B_CALIBRATION_FN_EDDY_SIM.csv"
after subtracting a piecewise linear function and normalizing, with different values of .
The masks are saved in the training and validation dataframes in
~/cernbox/hysteresis/dipole/notebooks/dynamic-downsampling/processed_dataframes/train_sim_df.parquet
~/cernbox/hysteresis/dipole/notebooks/dynamic-downsampling/processed_dataframes/val_sim_df.parquet
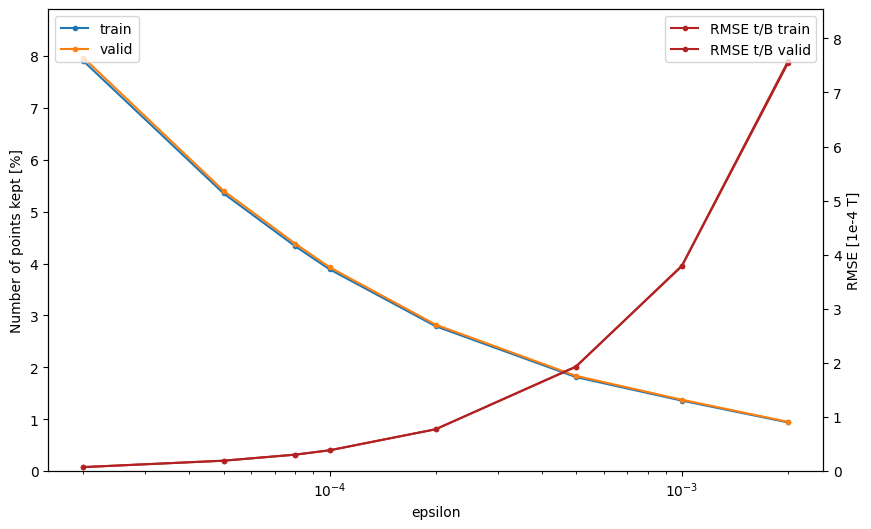 Using between and makes effective downsampling between 10 and 100, however with makes the RMSE increase drastically. Therefore the max that should be used is .
Using between and makes effective downsampling between 10 and 100, however with makes the RMSE increase drastically. Therefore the max that should be used is .
Preventing large time gaps
When large amounts of continuous points are removed, the time gap can be large, making outliers, that worsens the granularity of when normalized. Therefore there is need to re-add back points that are outside of .
Points are added evenly distributed with a random gap size where there are large time skips. This adds between and points, but reducing the maximum time value from 3000+ to below 500, which gives a time dynamic range of 0-500.
The following is from a jupyter notebook where timesteps are added:
Average time step size
For the optimal , the average median step is 36 ms. With a sequence length of 600 this equates to 21s of data on average.
With a context sequence length of 1200 and target sequence length of 600 we get
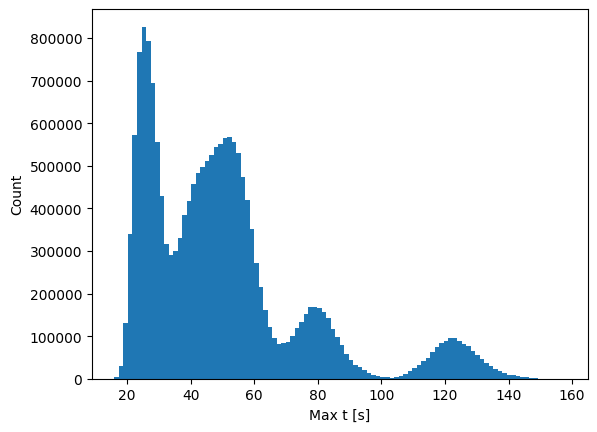
distribution